Cancer Mutation To Drug
A web server for anti-cancer drugs prediction based on gene mutation.
A web server for anti-cancer drugs prediction based on gene mutation.

Step 1: Enter your interest genes and mutations in the search box

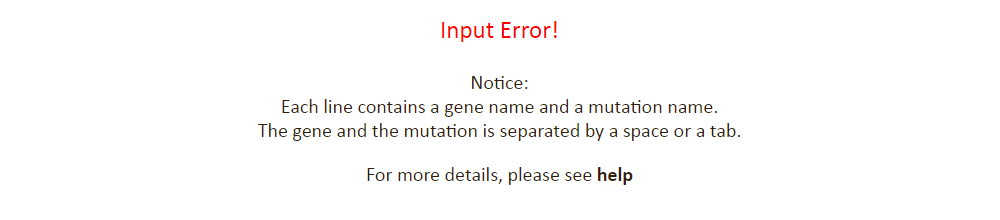
Notice:
Each line contains a gene name and a mutation name.
The gene and the mutation is separated by a space or a tab.
We recommend that you use official gene symbols and refSNP number IDs as input parameters.
Human official gene symbol can be found in GeneCards. It contains all annotated and predicted human genes.
The refSNP number ID can be found in the Single Nucleotide Polymorphism Database (dbSNP).
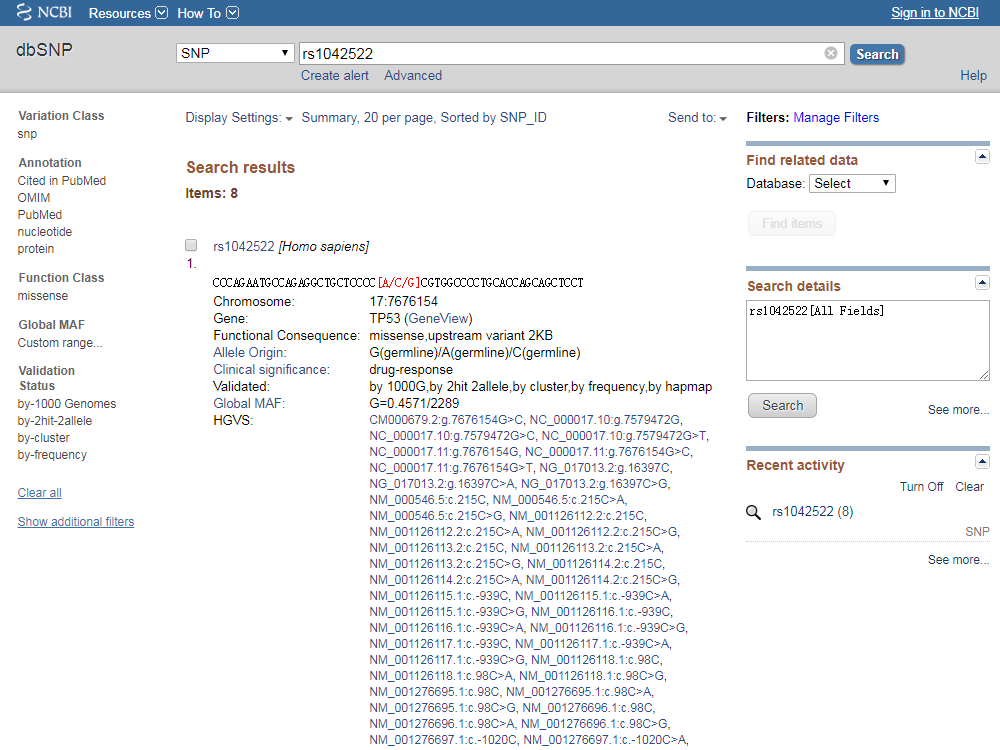
The dbSNP contains a range of molecular variation: (1) single nucleotide polymorphism (SNPs), (2) short deletion and insertion polymorphisms (indels/DIPs), (3) microsatellite markers or short tandem repeats (STRs), (4) multinucleotide polymorphisms (MNPs), (5) heterozygous sequences, and (6) named variants. The following is an example of refSNP number ID.
Step 2. Run Cancer Mutation To Drug and wait for few miniutes
Click "Predict" button to run Anti-Bacteria Predictor.

If input correctly, the page will jump to the wait page. In the wait page, you can copy the query ID, the query ID will be used when outputting the result.

If input error, the page will display an error message. You need to check the input genes and mutations and resubmit.
Step 3: Get and download your results
The wait page will automatically jump into the result page after some time.
You can also use the query ID to get the results. Paste your query ID to the web link:
http://drug.kiz.ac.cn:8080/CMTD/result/?q=Your query ID
Then paste the link into the address bar of the browser.
In the result page, the topleft box shows the input compound and the topright box shows a brief description of the results and a download link.
The detailed results including the following items:
# Rank = Ranked by FDR value.
# Drug = DrugBank ID and structure of the predicted compounds.
# Status = Current state of the drug (Approved, Experimental, Investigational, Withdraw, etc.).
# Target = Target proteins of the predicted compounds.
# P Value = Hypergeometric test P value. A P value less than 0.05 is considered significant.
# FDR = False discovery rate corrected P value. A FDR value less than 0.05 is considered significant.
You can also download the results by click the "Download" link.
